About AMICI
AMICI provides a Python and C++ interface to the SUNDIALS solvers CVODES (for ODEs) and IDAS (for DAEs). AMICI allows the user to read differential equation models specified as SBML or PySB and automatically compiles such models into Python modules or C++ libraries. All necessary functions are transformed into native C++ code, which allows for efficient simulation.
Beyond forward integration, the compiled simulation file also allows for forward sensitivity analysis, steady state sensitivity analysis and adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient computation in parameter estimation of biochemical reaction models but it is also applicable to a wider range of differential equation constrained optimization problems.
Features
SBML import
PySB import
Generation of C++ code for model simulation and sensitivity computation
Access to and high customizability of CVODES and IDAS solver
Python and C++ interface
Sensitivity analysis (first-order)
forward
steady state
adjoint
Pre-equilibration and pre-simulation conditions
Support for discrete events and logical operations
Interfaces & workflow
The AMICI workflow starts with importing a model from either SBML (Python) or PySB (Python). From this input, all equations for model simulation are derived symbolically and C++ code is generated. This code is then compiled into a plain C++ library or a Python module, and is then used for model simulation.
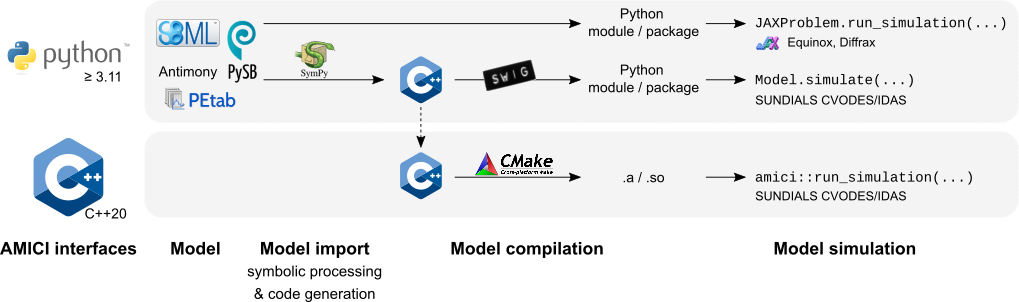
The functionality of the Python and C++ interfaces differ, as shown in the following table:
Feature \ Interface |
Python |
C++ |
|---|---|---|
SBML import |
yes (details) |
no |
PySB import |
yes |
no |
DAE import |
yes |
no |
Forward sensitivities |
yes |
yes |
Adjoint sensitivities |
yes |
yes |
Steadystate sensitivities |
yes |
yes |
Events |
yes |
yes |
yes |
yes |
|
yes |
yes |